Abstract
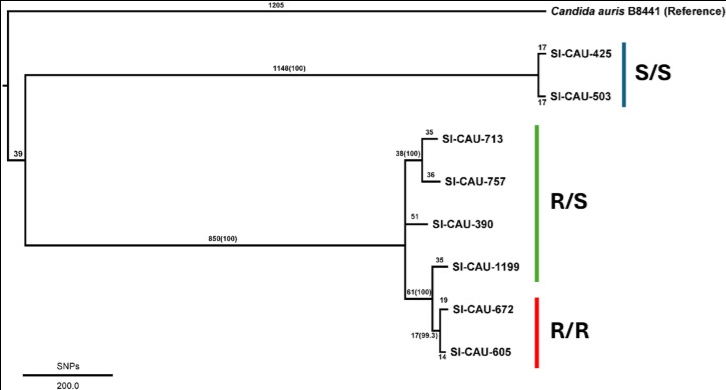
Candida auris, one of the emerging multidrug-resistant Candida species, was first discovered in 2009 from the ear canal of a patient in Tokyo, Japan. Since then, several studies exhibit the possible threats of C. auris due to its versatile resistance to frontline antifungal agents. The number of studies focusing on genetic variants associated with multidrug resistance has risen over a decade. In this study, we retrieved eight C. auris strains, originated from clinical isolates in Bangkok, Thailand, and sequenced all genomes using long-read oxford nanopore technology. We have identified genome-wide single nucleotide polymorphisms (SNPs) and found that the typical genetic variation, Y132F, in ERG11 was shared between fluconazole-resistant strains, as previously reported. The fluconazole-resistant strains also exclusively harboured other nonsynonymous SNPs suggesting new mutation candidates associated with fluconazole resistance. On the caspofungin resistance aspect, shared SNPs between caspofungin-resistant strains were also found in genes associated with various biological pathways, including G32E in CDC10, a gene encoding the protein related to cell wall integrity. The other mutation was found in fungal-specific transcription factors in the Zn (II)2-Cys6 binuclear cluster, which potentially plays a central role in regulating membrane efflux pumps or cell wall production. An amino acid substitution in FKS1, a well-known protein associated with echinocandin resistance, was found in only one of two caspofungin-resistant strains at the newly discovered position, I1361T. In short, the results of our study have confirmed the typical genetic variants underpinning multidrug resistance in C. auris strains and proposed the possible new pathways involved.