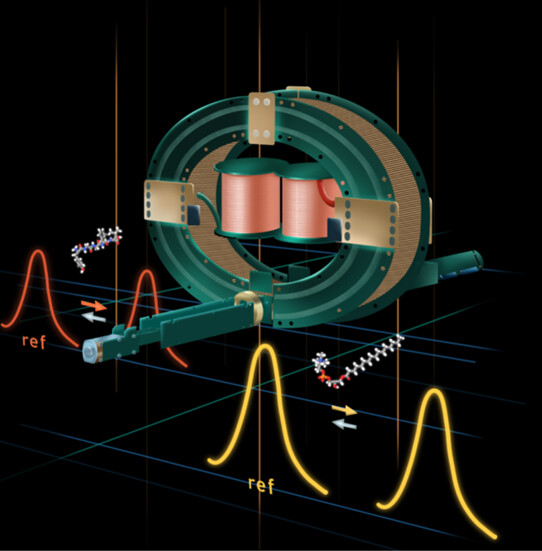
Abstract Direct-infusion mass spectrometry (DI-MS) and mass spectrometry imaging (MSI) are powerful techniques for lipidomics research. However, annotating isomeric and isobaric lipids with these methods is challenging due to the absence of chromatographic separation. Recently, cyclic ion mobility mass spectrometry (cIM-MS) has been proposed to overcome this limitation. However, fluctuations in room conditions can affect […]
Category Archives: Research Innovation
Abstract Introduction: To update the global burden of inflammatory bowel disease (IBD) using data from the Global Burden of Disease 2021. Methods: Data from Global Burden of Disease 2021 were analyzed to assess the IBD burden. Results: In 2021, there were 375,140 new cases and 3.83 million total cases of IBD. Elderly onset IBD accounted […]
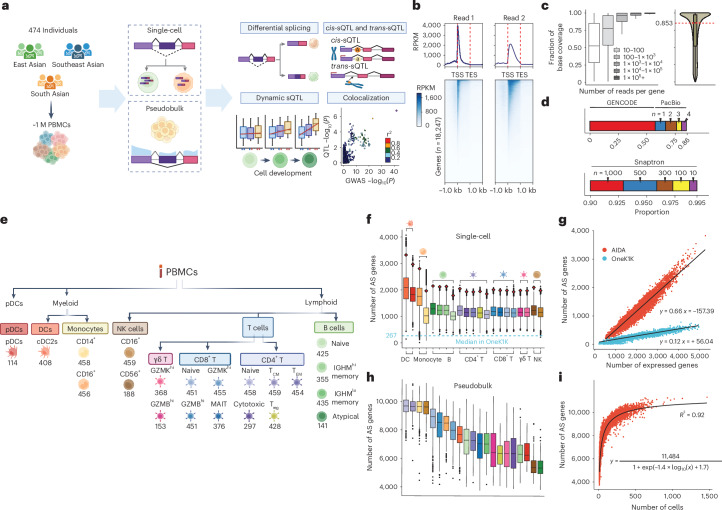
Abstract Alternative splicing contributes to complex traits, but whether this differs in trait-relevant cell types across diverse genetic ancestries is unclear. Here we describe cell-type-specific, sex-biased and ancestry-biased alternative splicing in ~1 M peripheral blood mononuclear cells from 474 healthy donors from the Asian Immune Diversity Atlas. We identify widespread sex-biased and ancestry-biased differential splicing, […]
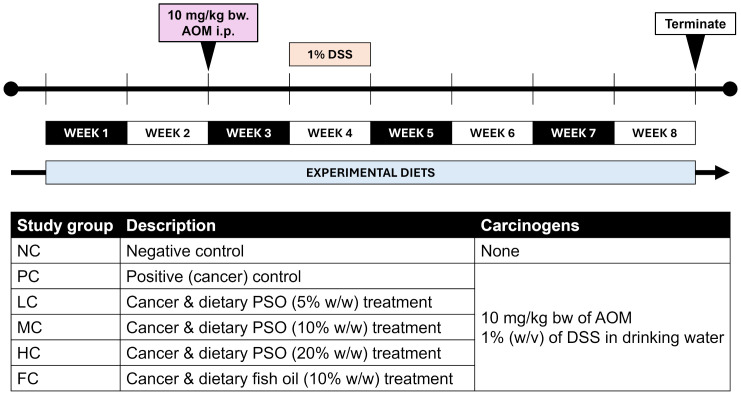
Abstract The present investigation explores into the influence of dietary nutrients, particularly alpha-linolenic acid (ALA), a plant-derived omega-3 fatty acid abundant in perilla seed oil (PSO), on the development of colitis-associated colorectal cancer (CRC). The study employs a mouse model to scrutinize the effects of ALA-rich PSO in the context of inflammation-driven CRC. Perilla seeds […]
Abstract Background The study of non-communicable diseases (NCDs) in a developing country like Thailand has rarely been conducted in long-term cohorts, especially among the working-age population. We aim to assess the prevalence and incidence of risk factors and their associations underlying NCDs, especially type-2 diabetes mellitus (T2DM) among healthcare workers enrolled in the Siriraj Health […]
Abstract Accurate taxonomic profiling of microbial taxa in a metagenomic sample is vital to gain insights into microbial ecology. Recent advancements in sequencing technologies have contributed tremendously toward understanding these microbes at species resolution through a whole shotgun metagenomic approach. In this study, we developed a new bioinformatics tool, coverage-based analysis for identification of microbiome […]
Abstract Vascular Ehlers–Danlos syndrome or Ehlers–Danlos syndrome type IV (vEDS) is a connective tissue disorder characterised by skin hyperextensibility, joint hypermobility and fatal vascular rupture caused by COL3A1 mutations that affect collagen III expression, homo-trimer assembly and secretion. Along with collagens I, II, V and XI, collagen III plays an important role in the extracellular […]
Abstract The revised WHO guidelines for classifying and grading brain tumors include several copy number variation (CNV) markers. The turnaround time for detecting CNVs and alterations throughout the entire genome is drastically reduced with the customized read incremental approach on the nanopore platform. However, this approach is challenging for non-bioinformaticians due to the need to […]
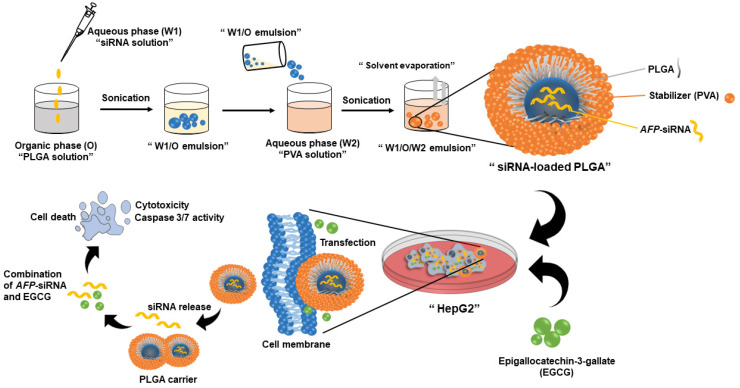
Abstract To develop a potential cancer treatment, we formulated a novel drug delivery platform made of poly(lactic-co-glycolic) acid (PLGA) and used a combination of an emerging siRNA technology and an extracted natural substance called catechins. The synthesized materials were characterized to determine their properties, including morphology, hydrodynamic size, charge, particle stability, and drug release profile. […]
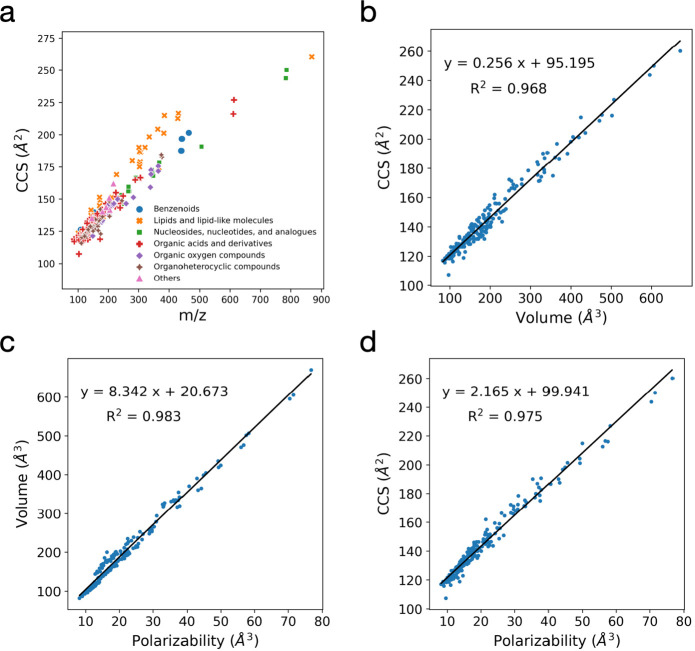
Abstract The rotationally averaged collision cross-section (CCS) determined by ion mobility-mass spectrometry (IM-MS) facilitates the identification of various biomolecules. Although machine learning (ML) models have recently emerged as a highly accurate approach for predicting CCS values, they rely on large data sets from various instruments, calibrants, and setups, which can introduce additional errors. In this […]