Abstract
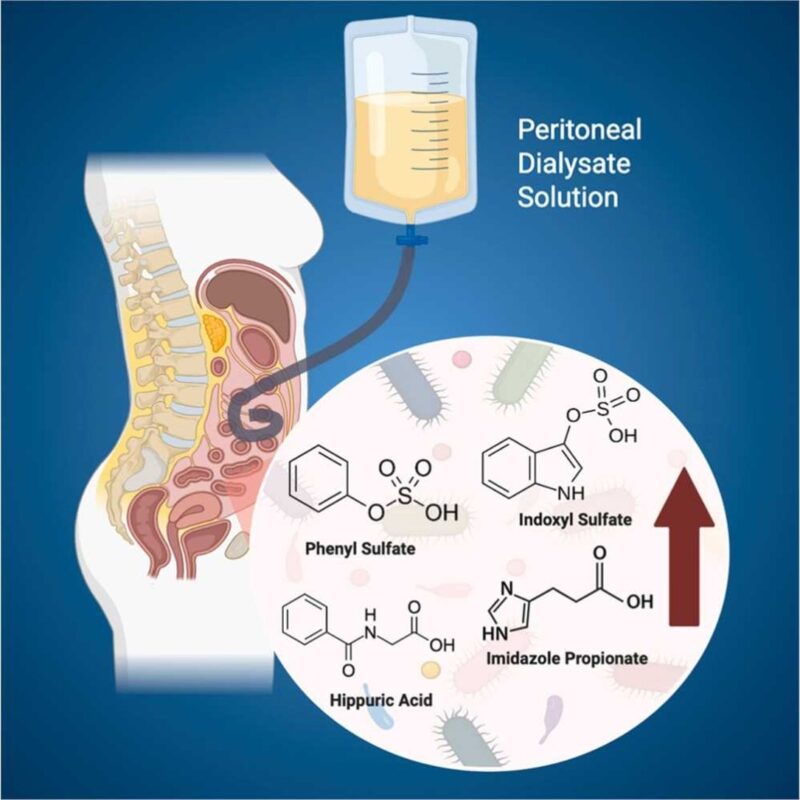
We developed a robust LC–MS/MS method for the simultaneous quantification of 16 uremic toxins (UTs) and 14 bile acids (BAs) in plasma and fecal samples within a single method. The method demonstrated high sensitivity, broad metabolite coverage, and excellent accuracy, precision, and throughput. Using this platform, targeted metabolites were quantified in peritoneal dialysis (PD) patients (n = 31) and healthy controls (HC; n = 60). Of the 30 targeted metabolites included in the validation method, 20 were detected in fecal samples and 12 in plasma in this study. Fecal samples exhibited greater BA diversity, whereas UTs were evenly distributed across both matrices. Fecal profiles showed minimal differences between PD and HC, suggesting limited gut-level alteration. In contrast, plasma analysis revealed nine metabolites significantly elevated in PD, including indoxyl sulfate, phenyl sulfate, hippuric acid, and imidazole propionate (ImP), lithocholic acid, cinnamoylglycine, m-hydroxyhippuric acid, phenylacetylglutamine, and phenylacetylglycine. Notably, plasma ImP—an underexplored metabolite—was elevated independently of diabetes or cardiovascular disease, implicating impaired renal clearance as its primary driver. These results highlight the systemic impact of gut-derived metabolites in kidney failure and position targeted UT–BA profiling as a powerful complementary tool for clinical metabolomics in chronic kidney disease and PD.
https://www.sciencedirect.com/science/article/pii/S2001037025005082